Published in the September 6, 2016 issue of Cell Reports, Standardized Whole-Blood Transcriptional Profiling Enables the Deconvolution of Complex Induced Immune Responses describes the use of the Myriad RBM TruCulture® platform for standardized ex vivo immune stimulation and a high-throughput method for gene expression profiling to explore immune variance within a population.
Standardized Whole-Blood Transcriptional Profiling Enables the Deconvolution of Complex Induced Immune Responses
Urrutia, AlejandraAbel, Laurent et al.
Cell Reports , Volume 16 , Issue 10 , 2777 – 2791
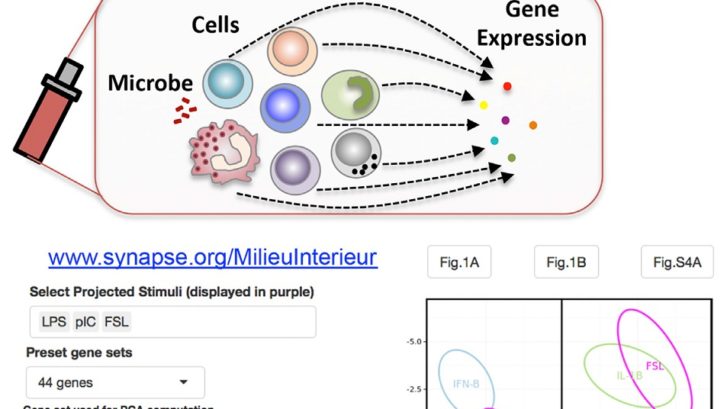
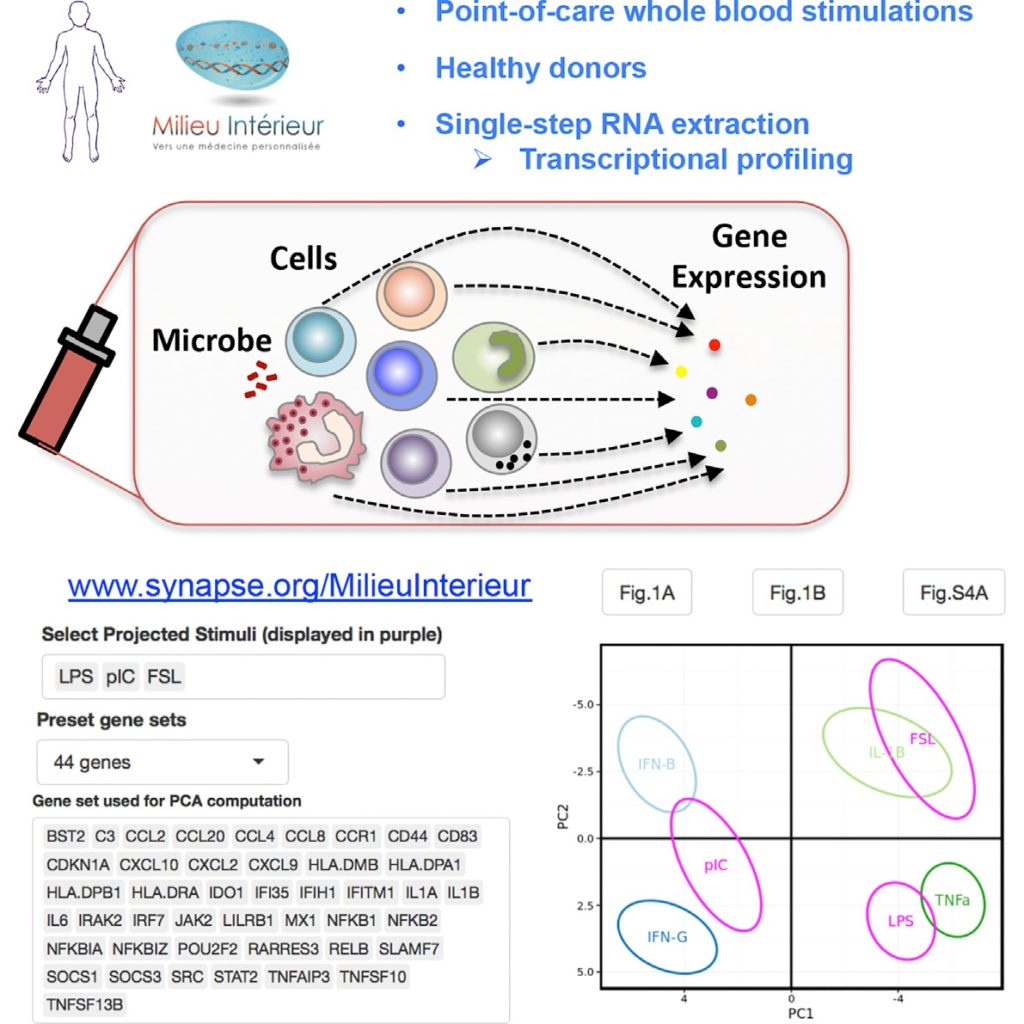
Urrutia, et al, utilized TruCulture tubes with 3 groups of stimuli for this study: purified cytokines, Toll-like receptor (TLR) agonists, and whole microbes. The study looked at the expression levels of different genes in 25 subjects following TruCulture incubation with twenty clinically relevant immune system stimulants. The research demonstrated that expression levels from as few as 44 of the 572 genes tested could segregate responses from the various stimuli and reveal variations in immune response.
“We are excited by the results of this study which demonstrated that a relatively small subset of gene expression targets could explain the variability in immune response to different immune stimuli seen among patients,” said Matthew Albert, co-coordinator of Milieu Interieur and director of the Immunobiology of Dendritic Cells Unit at the Institut Pasteur. “The ability of TruCulture to perform this analysis in an ex-vivo environment is truly unique making it an invaluable tool for assessing potential biomarkers relevant to immune response in patients.”
Data analysis revealed that each stimulus caused a specific pattern of gene expression and that variation between subjects can reveal new signaling pathways and feedback loop interactions. Neither type I (IFNa/b) nor type II (IFNg) interferon genes were expressed in response to any of the cytokine stimuli, but the IL-1b mRNA was highly induced by TNFa and IL-1b stimulation, indicating a positive feedback loop with IL-1b protein inducing IL-1b expression.
This study shows that using the TruCulture system for whole blood cultures coupled with single-step RNA extraction, minimizes pre-analytical bias and produces highly reproducible results, which is essential for multicenter population-based studies and clinical trials.
The Milieu Intérieur project coordinated by the Institut Pasteur, is a population-based study that aims to establish the determinates of a healthy immune response. To learn more about the Milieu Intérieur project and to review these results on a data visualization resource deployed in R-SHINY, visit the Milieu Intérieur website.
The objectives of the Milieu Intérieur project are to:
- Establish a deep understanding of immune variance;
- Define how immune phenotypic variation is genetically controlled and;
- Ascertain the role of the microbiota in regulating immune programs.
These efforts will establish parameters for stratifying individuals within a population, thus making it possible to glean meaningful interpretation from measurements of stress-induced host response. In achieving this goal, the project will provide a foundation for defining perturbations in an individual’s immune responses, thus laying the foundation for personalized medicine.